How hard is it to build your own top performance quantum circuit simulator? Does it really needs thousands of lines of code to implement it? At least in Julia language, you don’t! We can easily achieve top performance via a few hundreds of code while supporting CUDA and symbolic calculation.
Like my previous blog posts, you can do it in ONE DAY as well. I’ll introduce how to do this with Julia language while going through some common tricks for high performance computing in Julia language. I won’t talk much about the Julia language itself or it will be a very long blog post, thus if you want to follow this blog post but you don’t know how to use the Julia programming language yet, I would suggest you to checkout materials here first.
Table of contents
Open Table of contents
Background
Quantum computing has been a popular research topic in recent years. And building simulators can be useful for related research. I’m not going to give you a full introduction of what is quantum computing in this blog post, but I find you this nice tutorial from Microsoft if you are interested in knowing what is the quantum computing. And you don’t really need to understand everything about quantum computing to follow this blog post - the emulator itself is just about special matrix-vector or matrix-matrix multiplication.
So to be simple, simulating quantum circuits, or to be more specific simulating how quantum circuits act on a quantum register, is about how to calculate large matrix-vector multiplication that scales exponentially. The most brute-force and accurate way of doing it via full amplitude simulation, which means we do this matrix-vector multiplication directly.
The vector contains the so-called quantum state and the matrices are quantum gate, which are usually small. The diagram of quantum circuits is a representation of these matrix multiplications. For example, the X gate is just a small matrix
In theory, there is no way to simulate a general quantum circuit (more precisely, a universal gate set) efficiently, however, in practice, we could still do it within a rather small scale with some tricks that make use of the structure of the gates.
To know how to calculate a quantum circuit in the most naive way, we need to know two kinds of mathematical operations
Tensor Product/Kronecker Product, this is represented as two parallel lines in the quantum circuit diagram, e.g

and by definition, this can be calculated by
Matrix Multiplication, this is the most basic linear algebra operation, I’ll skip introducing this. In quantum circuit diagram, this is represented by blocks connected by lines.

As a conclusion of this section, you can see simulating how pure quantum circuits act on a given quantum state is about how to implement some special type of matrix-vector multiplication efficiently. If you know about BLAS (Basic Linear Algebra Subprograms), you will realize this kind of operations are only BLAS level 2 operations, which does not require any smart tiling technique and are mainly limited by memory bandwidth.
So let’s do it!
Implementing general unitary gate
Thus the simplest way of simulating a quantum circuit is very straightforward: we can just make use of Julia’s builtin functions:
kron and *.
using LinearAlgebra
function naive_broutine!(r::AbstractVector, U::AbstractMatrix, loc::Int)
n = Int(log2(length(r))) # get the number of qubits
return kron(I(1<<loc), U, I(1<<(n-loc-1))) * r
end
However, this is obviously very inefficient:
- we need to allocate a matrix every time we try to evaluate the gate.
- the length of the vector can only be , thus we should be able to calculate it faster with this knowledge.
I’ll start from the easiest thing: if we know an integer is , it is straight forward to find out by the following method
log2i(x::Int64) = !signbit(x) ? (63 - leading_zeros(x)) : throw(ErrorException("nonnegative expected ($x)"))
log2i(x::UInt64) = 63 - leading_zeros(x)
this is because we already know how long our integer is in the program by looking at its type, thus simply minus the number of leading zeros would give us the answer. But don’t forget to raise an error when it’s an signed integer type. We can make this work on any integer type by the following way
for N in [8, 16, 32, 64, 128]
T = Symbol(:Int, N)
UT = Symbol(:UInt, N)
@eval begin
log2i(x::$T) =
!signbit(x) ? ($(N - 1) - leading_zeros(x)) :
throw(ErrorException("nonnegative expected ($x)"))
log2i(x::$UT) = $(N - 1) - leading_zeros(x)
end
end
the command @eval here is called a macro in Julia programming language, it can be used to generate code. The above code generates the implementation of log2i for signed
and unsigned integer types from 8 bits to 128 bits.
Let’s now consider how to write the general unitary gate acting on given locations of qubits.
function broutine!(r::AbstractVector, U::AbstractMatrix, locs::NTuple{N, Int}) where N
end
this matrix will act on some certain qubits in the register, e.g given a 8x8 matrix we want it to act on the 1st, 4th and 5th qubit. Based on the implementation of X and Z we know this is about multiplying this matrix on the subspace of 1st, 4th and 5th qubit, which means we need to construct a set of new vectors whose indices iterate over the subspace of 0xx00x, 0xx01x, 0xx10x, 0xx11x etc. Thus the first thing we need to do is to find a generic way to iterate through the subspace of 0xx00x then by adding an offset such as 1<<1 to each index in this subspace, we can get the subspace of 0xx01x etc.
Iterate through the subspace
To iterate through the subspace, we could iterate through all indices in the subspace. For each index, we move each bit to its position in the whole space (from first bit to the last).
This will give us the first subspace which is 0xx00x.
Before we move on, I need to introduce the concept of binary masks: it is an integer that can help us “filter” out some binary values, e.g
we want to know if a given integer’s 4th and 5th bit, we can use a mask 0b11000, where its 4th and 5th bit are 1 the rest is 0, then we
can use an and operation get get the value. Given the location of bits, we can create a binary mask via the following bmask function
function bmask(itr)
isempty(itr) && return 0
ret = 0
for b in itr
ret += 1 << (b - 1)
end
return ret
end
where itr is some iterable. However there are quite a few cases that we don’t need to create it via a for-loop, so we can specialize this function
on the following types
function bmask(range::UnitRange{Int})
((1 << (range.stop - range.start + 1)) - 1) << (range.start - 1)
end
however, we maybe want to make the implementation more general for arbitrary integer types, so let’s use a type variable T!
function bmask(::Type{T}, itr) where {T<:Integer}
isempty(itr) && return 0
ret = zero(T)
for b in itr
ret += one(T) << (b - 1)
end
return ret
end
function bmask(::Type{T}, range::UnitRange{Int})::T where {T<:Integer}
((one(T) << (range.stop - range.start + 1)) - one(T)) << (range.start - 1)
end
However after we put a type variable as the first argument, it is not convenient when we just want to use Int64 anymore,
let’s create a few convenient methods then
bmask(args...) = bmask(Int, args...)
# this is for removing the infinity call of the later function
bmask(::Type{T}) where {T<:Integer} = zero(T)
bmask(::Type{T}, positions::Int...) where {T<:Integer} = bmask(T, positions)
The final implement would look like the following
bmask(args...) = bmask(Int, args...)
bmask(::Type{T}) where {T<:Integer} = zero(T)
bmask(::Type{T}, positions::Int...) where {T<:Integer} = bmask(T, positions)
function bmask(::Type{T}, itr) where {T<:Integer}
isempty(itr) && return 0
ret = zero(T)
for b in itr
ret += one(T) << (b - 1)
end
return ret
end
function bmask(::Type{T}, range::UnitRange{Int})::T where {T<:Integer}
((one(T) << (range.stop - range.start + 1)) - one(T)) << (range.start - 1)
end
To move the bits in subspace to the right position, we need to iterate through all the contiguous region in the bitstring, e.g for 0xx00x, we
move the 2nd and 3rd bit in subspace by 3 bits together, this can be achieved by using a bit mask 001 and the following binary operation
(xxx & ~0b001) << 1 + (xxx & 0b001) # = xx00x
we define this as a function called lmove:
@inline lmove(b::Int, mask::Int, k::Int)::Int = (b & ~mask) << k + (b & mask)
we mark this function @inline
here to make sure the compiler will always inline it,
now we need to generate all the masks by counting contiguous region of the given locations
function group_shift(locations)
masks = Int[]
region_lens = Int[]
k_prv = -1
for k in locations
# if current position in the contiguous region
# since these bits will be moved together with
# the first one, we don't need to generate a
# new mask
if k == k_prv + 1
region_lens[end] += 1
else
# we generate a bit mask where the 1st to k-th bits are 1
push!(masks, bmask(0:k-1))
push!(region_lens, 1)
end
k_prv = k
end
return masks, region_lens
end
now to get the index in the whole space, we simply move each contiguous region by the length of their region,
for s in 1:n_regions
index = lmove(index, masks[s], region_lens[s])
end
where the initial value of index is the subspace index, and after the loop, we will get the index in the whole space.
Now, since we need to iterate the all the possible indices, it would be very convenient to have an iterator, let’s implement this as an iterator,
struct BitSubspace
n::Int # total number of bits
n_subspace::Int # number of bits in the subspace
masks::Vector{Int} # masks
region_lens::Vector{Int} # length of each region
end
and we can construct it via
function BitSubspace(n::Int, locations)
masks, region_lens = group_shift(locations)
BitSubspace(1 << (n - length(locations)), length(masks), masks, region_lens)
end
now, let’s consider the corresponding whole-space index of each index in the subspace.
@inline function Base.getindex(it::BitSubspace, i)
index = i - 1
for s in 1:it.n_subspace
@inbounds index = lmove(index, it.masks[s], it.region_lens[s])
end
return index
end
now let’s overload some methods to make this object become an iterable object
Base.length(it::BitSubspace) = it.n
Base.eltype(::BitSubspace) = Int
@inline function Base.iterate(it::BitSubspace, st = 1)
if st > length(it)
return nothing
else
return it[st], st + 1
end
end
let’s try it! it works!
julia> for each in BitSubspace(5, [1, 3, 4])
println(string(each, base=2, pad=7))
end
00000
00010
10000
10010
Multiply matrix in subspace
now we know how to generate the indices in a subspace, we need to multiply the matrix to each subspace,
e.g for a unitary on the 1, 3, 4 qubits of a 5-qubit register, we need to multiply the matrix at 0xx0x,
0xx1x, 1xx0x and 1xx1x. Thus we can create the subspace of x00x0 by BitSubspace(5, [1, 3, 4])
and subspace of 0xx0x by BitSubspace(5, [2, 5]), then add each index in x00x0 to 0xx0x, which looks like
subspace1 = BitSubspace(5, [1, 3, 4])
subspace2 = BitSubspace(5, [2, 5])
# Julia uses 1-based index, we need to convert it
indices = collect(b + 1 for b in subspace2)
@inbounds for i in subspace1
# add an offset i to all the indices of 0xx0x
# this will give us 0xx0x, 0xx1x, 1xx0x, 1xx1x
idx = indices .+ i
state[idx] = U * state[idx] # matrix multiplication on the subspace
end
now we notice subspace2 is the complement subspace of subspace1 because the full space if [1, 2, 3, 4, 5], so let’s redefine our BitSubspace
constructor a bit, now instead of define the constructor BitSubspace(n, locations) we define two functions to create this object bsubspace(n, locations) and
bcomspace(n, locations) which stands for binary subspace and binary complement space, the function bsubspace will create subspace1 and the function
bcomspace(n, locations) will create subspace2.
They have some overlapping operations, so I move them to an internal function _group_shift
@inline function group_shift(locations)
masks = Int[]
shift_len = Int[]
k_prv = -1
for k in locations
_group_shift(masks, shift_len, k, k_prv)
k_prv = k
end
return masks, shift_len
end
@inline function complement_group_shift(n::Int, locations)
masks = Int[]
shift_len = Int[]
k_prv = -1
for k in 1:n
k in locations && continue
_group_shift(masks, shift_len, k, k_prv)
k_prv = k
end
return masks, shift_len
end
@inline function _group_shift(masks::Vector{Int}, shift_len::Vector{Int}, k::Int, k_prv::Int)
# if current position in the contiguous region
# since these bits will be moved together with
# the first one, we don't need to generate a
# new mask
if k == k_prv + 1
shift_len[end] += 1
else
# we generate a bit mask where the 1st to k-th bits are 1
push!(masks, bmask(0:k-1))
push!(shift_len, 1)
end
end
thus our routine will look like the following
function broutine!(st::AbstractVector, U::AbstractMatrix, locs::NTuple{N, Int}) where N
n = log2dim1(st)
subspace = bsubspace(n, locs)
comspace = bcomspace(n, locs)
indices = [idx + 1 for idx in comspace]
@inbounds @views for k in subspace
idx = indices .+ k
st[idx] = U * st[idx]
end
return st
end
the log2dim1 is just a convenient one-liner log2dim1(x) = log2i(size(x, 1)). And we use @inbounds here to tell the Julia compiler
that we are pretty sure all our indices are inbounds! And use @views to tell Julia we are confident at mutating our arrays so please
use a view and don’t allocate any memory!
Now you may notice: the iteration in our implementation is independent and may be reordered! This means we can easily make this parallel. The simplest way to parallelize it is via multi-threading. In Julia, this is extremely simple,
function threaded_broutine!(st::AbstractVector, U::AbstractMatrix, locs::NTuple{N, Int}) where N
n = log2dim1(st)
subspace = bsubspace(n, locs)
comspace = bcomspace(n, locs)
indices = [idx + 1 for idx in comspace]
@inbounds @views Threads.@threads for k in subspace
idx = indices .+ k
st[idx] = U * st[idx]
end
return st
end
but wait, this will give you en error MethodError: no method matching firstindex(::BitSubspace), this is simply because
the @threads wants calculate which indices it needs to put into one thread using firstindex, so let’s define it
Base.firstindex(::BitSubspace) = 1
thus the final implementation of subspace would looks like the following
@inline function _group_shift(masks::Vector{Int}, shift_len::Vector{Int}, k::Int, k_prv::Int)
# if current position in the contiguous region
# since these bits will be moved together with
# the first one, we don't need to generate a
# new mask
if k == k_prv + 1
shift_len[end] += 1
else
# we generate a bit mask where the 1st to k-th bits are 1
push!(masks, bmask(0:k-1))
push!(shift_len, 1)
end
end
@inline function group_shift(locations)
masks = Int[]
shift_len = Int[]
k_prv = -1
for k in locations
_group_shift(masks, shift_len, k, k_prv)
k_prv = k
end
return masks, shift_len
end
@inline function complement_group_shift(n::Int, locations)
masks = Int[]
shift_len = Int[]
k_prv = -1
for k in 1:n
k in locations && continue
_group_shift(masks, shift_len, k, k_prv)
k_prv = k
end
return masks, shift_len
end
struct BitSubspace
n::Int # number of bits in fullspace
sz_subspace::Int # size of the subspace
n_shifts::Int # number of shifts
masks::Vector{Int} # shift masks
shift_len::Vector{Int} # length of each shift
end
function Base.getindex(s::BitSubspace, i::Int)
index = i - 1
@inbounds for k in 1:s.n_shifts
index = lmove(index, s.masks[k], s.shift_len[k])
end
return index
end
Base.firstindex(s::BitSubspace) = 1
Base.lastindex(s::BitSubspace) = s.sz_subspace
Base.length(s::BitSubspace) = s.sz_subspace
Base.eltype(::BitSubspace) = Int
function Base.iterate(s::BitSubspace, st::Int = 1)
st <= length(s) || return
return s[st], st + 1
end
function bsubspace(n::Int, locs)
@assert issorted(locs)
masks, shift_len = group_shift(locs)
BitSubspace(n, 1 << (n - length(locs)), length(masks), masks, shift_len)
end
function bcomspace(n::Int, locs)
@assert issorted(locs)
masks, shift_len = complement_group_shift(n, locs)
BitSubspace(n, 1 << length(locs), length(masks), masks, shift_len)
end
function broutine!(st::AbstractVector, U::AbstractMatrix, locs::NTuple{N, Int}) where N
n = log2dim1(st)
subspace = bsubspace(n, locs)
comspace = bcomspace(n, locs)
indices = [idx + 1 for idx in comspace]
@inbounds @views for k in subspace
idx = indices .+ k
st[idx] = U * st[idx]
end
return st
end
here I changed the definition of struct BitSubspace to store the number of bits in fullspace so that we can print it nicely
function Base.show(io::IO, ::MIME"text/plain", s::BitSubspace)
indent = get(io, :indent, 0)
println(io, " "^indent, s.sz_subspace, "-element BitSubspace:")
if s.sz_subspace < 5
for k in 1:s.sz_subspace
print(io, " "^(indent+1), string(s[k]; base=2, pad=s.n))
if k != s.sz_subspace
println(io)
end
end
else # never print more than 4 elements
println(io, " "^(indent+1), string(s[1]; base=2, pad=s.n))
println(io, " "^(indent+1), string(s[2]; base=2, pad=s.n))
println(io, " "^(indent+1), "⋮")
println(io, " "^(indent+1), string(s[end-1]; base=2, pad=s.n))
print(io, " "^(indent+1), string(s[end]; base=2, pad=s.n))
end
end
let’s try it!
julia> bsubspace(5, (2, 3))
8-element BitSubspace:
00000
00001
⋮
11000
11001
julia> bcomspace(5, (2, 3))
4-element BitSubspace:
00000
00010
00100
00110
Implement controlled gate
Now I have introduced all the tricks for normal quantum gates, however, there are another important set of gates which is controlled gates. There are no new tricks, but we will need to generalize the implementation above a little bit.
General controlled unitary gate
Controlled unitary gate basically means when we see an index, e.g 010011, except applying our unitary matrix on the given location (e.g 1 and 2), we need to look
at the control qubit, if the control qubit is 0, we do nothing, if the control qubit is 1 we apply the matrix. (for inverse control gate, this is opposite)
Thus, this means the subspace we will be looking at contains 2 parts: the bits on control locations are 1 and the bits on gate locations are 0. We can define our
offset as following:
ctrl_offset(locs, configs) = bmask(locs[i] for (i, u) in enumerate(configs) if u != 0)
and the corresponding routine becomes
function routine!(st::AbstractVector, U::AbstractMatrix, locs::NTuple{N, Int}, ctrl_locs::NTuple{M, Int}, ctrl_configs::NTuple{M, Int}) where {N, M}
n = log2dim1(st)
subspace = bsubspace(n, sort([locs..., ctrl_locs...]))
comspace = bcomspace(n, locs)
offset = ctrl_offset(ctrl_locs, ctrl_configs)
indices = [idx + 1 for idx in comspace]
@inbounds @views for k in subspace
idx = indices .+ k .+ offset
st[idx] = U * st[idx]
end
return st
end
Optimize performance for small matrices
In most cases, the matrices of unitary gates we want to simulate are usually very small. They are usually of size 2x2 (1 qubit),
4x4 (2 qubit) or 8x8 (3 qubit). In these cases, we can consider using the StaticArray which is much faster than openBLAS/MKL for
small matrices, but we don’t need to change our routine! implementation, since Julia will specialize our generic functions
automatically:
using BenchmarkTools, StaticArrays
U1 = rand(ComplexF64, 8, 8);
U2 = @SMatrix rand(ComplexF64, 8, 8);
locs = (4, 9, 10);
st = rand(ComplexF64, 1<<15);
and we can see the benchmark
julia> @benchmark broutine!(r, $U1, $locs) setup=(r=copy($st))
BenchmarkTools.Trial:
memory estimate: 1.38 MiB
allocs estimate: 8201
--------------
minimum time: 489.581 μs (0.00% GC)
median time: 513.550 μs (0.00% GC)
mean time: 539.640 μs (4.09% GC)
maximum time: 1.403 ms (62.67% GC)
--------------
samples: 8451
evals/sample: 1
julia> @benchmark broutine!(r, $U2, $locs) setup=(r=copy($st))
BenchmarkTools.Trial:
memory estimate: 576.64 KiB
allocs estimate: 4105
--------------
minimum time: 182.967 μs (0.00% GC)
median time: 188.346 μs (0.00% GC)
mean time: 202.701 μs (6.45% GC)
maximum time: 999.731 μs (80.77% GC)
--------------
samples: 10000
evals/sample: 1
Using StaticArray is already very fast, But this is still space to optimize it, and because StaticArray will
store everything as a type in compile time, this will force us to compile things at runtime, which can make the first
time execution slow (since Julia uses just-in-time compilation,
it can specialize functions at runtime). Before we move forward, let me formalize the problem a bit more:
Now as you might have noticed, what we have been doing is implementing a matrix-vector multiplication but in subspace,
and we always know the indices inside the complement space, we just need to calculate its value in the full space, and
because of the control gates, we may need to add an offset to the indices in full space, but it is 0 by default,
thus this operation can be defined as following
function subspace_mul!(st::AbstractVector{T}, comspace, U, subspace, offset=0) where T
end
now let’s implement this in a plain for loop, if you happened to forget how to calculate matrix-vector multiplication,
an einstein summation notation may help:
where is a matrix and are indices in our subspace. Now we can write down our subspace multiplication function
function subspace_mul!(st::AbstractVector{T}, comspace, U, subspace, offset=0) where T
indices = [idx + 1 for idx in comspace]
y = similar(st, (size(U, 1), ))
idx = similar(indices)
@inbounds for k in subspace
for i in 1:size(U, 1)
idx[i] = indices[i] + k + offset
end
for i in 1:size(U, 1)
y[i] = zero(T)
for j in 1:size(U, 2)
y[i] += U[i, j] * st[idx[j]]
end
end
for i in 1:size(U, 1)
st[idx[i]] = y[i]
end
end
return st
end
if you are familiar with BLAS functions, there is a small difference with gemv routine: because we are doing multiplication
in a large space, we need to allocate a small vector to store intermediate result in the subspace and then assign the intermediate
result to the full space vector.
Now let’s use this implementation in our broutine! function:
function broutine!(st::AbstractVector, U::AbstractMatrix, locs::NTuple{N, Int}) where N
n = log2dim1(st)
subspace = bsubspace(n, locs)
comspace = bcomspace(n, locs)
subspace_mul!(st, comspace, U, subspace)
return st
end
function broutine!(st::AbstractVector, U::AbstractMatrix, locs::NTuple{N, Int}, ctrl_locs::NTuple{M, Int}, ctrl_configs::NTuple{M, Int}) where {N, M}
n = log2dim1(st)
subspace = bsubspace(n, sort([locs..., ctrl_locs...]))
comspace = bcomspace(n, locs)
offset = ctrl_offset(ctrl_locs, ctrl_configs)
subspace_mul!(st, comspace, U, subspace, offset)
return st
end
As you can see, it is faster now, but still slower than StaticArrays, this is because our compiler still has no access to the shape information
of your matrix
julia> @benchmark broutine!(r, $U1, $locs) setup=(r=copy($st))
BenchmarkTools.Trial:
memory estimate: 1008 bytes
allocs estimate: 11
--------------
minimum time: 247.516 μs (0.00% GC)
median time: 282.016 μs (0.00% GC)
mean time: 281.811 μs (0.00% GC)
maximum time: 489.902 μs (0.00% GC)
--------------
samples: 10000
evals/sample: 1
A direct observation is that the inner loop has a very small size in the case of quantum gates
for i in 1:size(U, 1)
y[i] = zero(T)
for j in 1:size(U, 2)
y[i] += U[i, j] * st[idx[j]]
end
end
if U is a 2x2 matrix, this can be written as
T1 = U[1, 1] * st[idx[1]] + U[1, 2] * st[idx[2]]
T2 = U[2, 1] * st[idx[1]] + U[2, 2] * st[idx[2]]
first you will find we don’t need our intermediate array y anymore! And moreover, notice that the order of T1 and T2 doesn’t matter
for this calculation, which means in principal they can be executed in parallel! But this is an inner loop, we don’t want to waste our
multi-thread resources to parallel it, instead we hope we can have SIMD. However, we don’t have to
call SIMD instructions explicitly, because in fact the compiler
can figure out how to use SIMD instructions for the 2x2 case itself, since it’s very obvious, and also because we have implicitly implied that we only
have a matrix of shape 2x2 by expanding the loop. So let’s just trust our compiler
function subspace_mul2x2!(st::AbstractVector{T}, comspace, U, subspace, offset=0) where T
indices_1 = comspace[1] + 1
indices_2 = comspace[2] + 1
@inbounds for k in subspace
idx_1 = indices_1 + k + offset
idx_2 = indices_2 + k + offset
T1 = U[1, 1] * st[idx_1] + U[1, 2] * st[idx_2]
T2 = U[2, 1] * st[idx_1] + U[2, 2] * st[idx_2]
st[idx_1] = T1
st[idx_2] = T2
end
return st
end
we can do similar things for 4x4 and 8x8 matrices, implementing them is quite mechanical, thus we will seek some macro magic
now
function subspace_mul4x4!(st::AbstractVector{T}, comspace, U, subspace, offset=0) where T
Base.Cartesian.@nextract 4 indices i -> comspace[i] + 1
Base.Cartesian.@nextract 4 U i->begin
Base.Cartesian.@nextract 4 U_i j->U[i, j]
end
for k in subspace
Base.Cartesian.@nextract 4 idx i-> k + indices_i + offset
Base.Cartesian.@nexprs 4 i -> begin
y_i = zero(T)
Base.Cartesian.@nexprs 4 j -> begin
y_i += U_i_j * st[idx_j]
end
end
Base.Cartesian.@nexprs 4 i -> begin
st[idx_i] = y_i
end
end
return st
end
function subspace_mul8x8!(st::AbstractVector{T}, comspace, U, subspace, offset=0) where T
Base.Cartesian.@nextract 8 indices i -> comspace[i] + 1
Base.Cartesian.@nextract 8 U i->begin
Base.Cartesian.@nextract 8 U_i j->U[i, j]
end
@inbounds for k in subspace
Base.Cartesian.@nextract 8 idx i-> k + indices_i + offset
Base.Cartesian.@nexprs 8 i -> begin
y_i = zero(T)
Base.Cartesian.@nexprs 8 j -> begin
y_i += U_i_j * st[idx_j]
end
end
Base.Cartesian.@nexprs 8 i -> begin
st[idx_i] = y_i
end
end
return st
end
In Julia the macro Base.Cartesian.@nextract will generate a bunch of variables like indices_1, indice_2 etc.
automatically at compile time for us, so we don’t need to do it ourselves. And then we can use Base.Cartesian.@nexprs
to implement the matrix multiplication statements and assign the values back to full space vector st. If you have questions
about how to use Base.Cartesian.@nextract and Base.Cartesian.@nexprs you can use the help mode in Julia REPL to check their
documentation. Now we will want to dispatch the method subspace_mul! to these specialized methods when we have a 2x2, 4x4
or 8x8 matrix, so we move our original plain-loop version subspace_mul! to a new function subspace_mul_generic!,
and dispatch methods based on the matrix size
function subspace_mul!(st::AbstractVector{T}, comspace, U, subspace, offset=0) where T
if size(U, 1) == 2
subspace_mul2x2!(st, comspace, U, subspace, offset)
elseif size(U, 1) == 4
subspace_mul4x4!(st, comspace, U, subspace, offset)
elseif size(U, 1) == 8
subspace_mul8x8!(st, comspace, U, subspace, offset)
else
subspace_mul_generic!(st, comspace, U, subspace, offset)
end
return st
end
if we try it on our previous benchmark, we will see we are faster than StaticArrays now!
julia> @benchmark broutine!(r, $U1, $locs) setup=(r=copy($st))
BenchmarkTools.Trial:
memory estimate: 512 bytes
allocs estimate: 8
--------------
minimum time: 141.577 μs (0.00% GC)
median time: 145.168 μs (0.00% GC)
mean time: 145.998 μs (0.00% GC)
maximum time: 169.246 μs (0.00% GC)
--------------
samples: 10000
evals/sample: 1
now since most of the quantum gates are 2x2 matrices, we will focus more on this case, recall that in the 2x2 matrix case,
there is only one location to specify, this will allow us to directly iterate through the subspace by adding up 2^loc, where
the variable loc is the integer represents the location of this gate. This will get us rid of all the heavier BitSubspace struct.
function broutine2x2!(st::AbstractVector{T}, U::AbstractMatrix, locs::Tuple{Int}) where T
U11 = U[1, 1]; U12 = U[1, 2];
U21 = U[2, 1]; U22 = U[2, 2];
step_1 = 1 << (first(locs) - 1)
step_2 = 1 << first(locs)
@inbounds for j in 0:step_2:size(st, 1)-step_1
for i in j+1:j+step_1
ST1 = U11 * st[i] + U12 * st[i + step_1]
ST2 = U21 * st[i] + U22 * st[i + step_1]
st[i] = ST1
st[i + step_1] = ST2
end
end
return st
end
let’s compare this and subspace_mul2x2!, to be fair we will directly call broutine! and it will call subspace_mul! then dispatch to subspace_mul2x2!.
julia> U = rand(ComplexF64, 2, 2);
julia> locs = (3, );
julia> st = rand(ComplexF64, 1<<15);
julia> @benchmark broutine!(r, $U, $locs) setup=(r=copy($st))
BenchmarkTools.Trial:
memory estimate: 512 bytes
allocs estimate: 8
--------------
minimum time: 67.639 μs (0.00% GC)
median time: 81.669 μs (0.00% GC)
mean time: 86.487 μs (0.00% GC)
maximum time: 125.038 μs (0.00% GC)
--------------
samples: 10000
evals/sample: 1
julia> @benchmark broutine2x2!(r, $U, $locs) setup=(r=copy($st))
BenchmarkTools.Trial:
memory estimate: 0 bytes
allocs estimate: 0
--------------
minimum time: 63.419 μs (0.00% GC)
median time: 64.369 μs (0.00% GC)
mean time: 64.757 μs (0.00% GC)
maximum time: 86.489 μs (0.00% GC)
--------------
samples: 10000
evals/sample: 1
this is only a little bit faster. Hmm, this is not very ideal, but notice that because step_1 can
be very small and it is an inner loop, we can then unroll this loop as long as it is small, so we can
now manually write
function broutine2x2!(st::AbstractVector{T}, U::AbstractMatrix, locs::Tuple{Int}) where T
U11 = U[1, 1]; U12 = U[1, 2];
U21 = U[2, 1]; U22 = U[2, 2];
step_1 = 1 << (first(locs) - 1)
step_2 = 1 << first(locs)
@inbounds if step_1 == 1
for j in 0:step_2:size(st, 1)-step_1
ST1 = U11 * st[j + 1] + U12 * st[j + 1 + step_1]
ST2 = U21 * st[j + 1] + U22 * st[j + 1 + step_1]
st[j + 1] = ST1
st[j + 1 + step_1] = ST2
end
elseif step_1 == 2
for j in 0:step_2:size(st, 1)-step_1
Base.Cartesian.@nexprs 2 i->begin
ST1 = U11 * st[j + i] + U12 * st[j + i + step_1]
ST2 = U21 * st[j + i] + U22 * st[j + i + step_1]
st[j + i] = ST1
st[j + i + step_1] = ST2
end
end
elseif step_1 == 4
for j in 0:step_2:size(st, 1)-step_1
Base.Cartesian.@nexprs 4 i->begin
ST1 = U11 * st[j + i] + U12 * st[j + i + step_1]
ST2 = U21 * st[j + i] + U22 * st[j + i + step_1]
st[j + i] = ST1
st[j + i + step_1] = ST2
end
end
elseif step_1 == 8
for j in 0:step_2:size(st, 1)-step_1
Base.Cartesian.@nexprs 8 i->begin
ST1 = U11 * st[j + i] + U12 * st[j + i + step_1]
ST2 = U21 * st[j + i] + U22 * st[j + i + step_1]
st[j + i] = ST1
st[j + i + step_1] = ST2
end
end
else
for j in 0:step_2:size(st, 1)-step_1
for i in j:8:j+step_1-1
Base.Cartesian.@nexprs 8 k->begin
ST1 = U11 * st[i + k] + U12 * st[i + step_1 + k]
ST2 = U21 * st[i + k] + U22 * st[i + step_1 + k]
st[i + k] = ST1
st[i + step_1 + k] = ST2
end
end
end
end
return st
end
the last loop is also partially unrolled by slicing our iteration range.
julia> @benchmark broutine2x2!(r, $U, $locs) setup=(r=copy($st))
BenchmarkTools.Trial:
memory estimate: 0 bytes
allocs estimate: 0
--------------
minimum time: 21.420 μs (0.00% GC)
median time: 21.670 μs (0.00% GC)
mean time: 21.818 μs (0.00% GC)
maximum time: 45.829 μs (0.00% GC)
--------------
samples: 10000
evals/sample: 1
this is now much faster than subspace_mul2x2!, as you see, by slightly change the abstraction
we implement, we exposed a small loop that can be unrolled! So let’s delete our subspace_mul2x2!
and use this method instead:
function broutine!(st::AbstractVector, U::AbstractMatrix, locs::NTuple{N, Int}) where N
size(U, 1) == 2 && return broutine!(st, U, locs)
n = log2dim1(st)
subspace = bsubspace(n, locs)
comspace = bcomspace(n, locs)
subspace_mul!(st, comspace, U, subspace)
return st
end
now let’s think about how to unroll the small matrix for the controlled gate case: the term controlled gate simply means
when we see there is 1 (or 0 for inverse control) at the control location, we apply the matrix in subspace, or we don’t.
so we can just check the control location’s configuration inside the loop, to do this we can create two masks: a control
location mask ctrl_mask and a control flag mask flag_mask
ctrl_mask = bmask(ctrl_locs)
flag_mask = reduce(+, 1 << (ctrl_locs[i] - 1) for i in 1:length(ctrl_locs) if ctrl_configs[i])
then we just need to check the bits on ctrl_locs to see if they are the same with flag_mask, we can implement a function
ismatch to do this
ismatch(index::T, mask::T, target::T) where {T<:Integer} = (index & mask) == target
thus the implementation will look very similar to the un-controlled one, although it is evil to copy-past, to be able to implement it within a day, I’ll just do so
function broutine2x2!(st::AbstractVector, U::AbstractMatrix, locs::Tuple{Int}, ctrl_locs::NTuple{M, Int}, ctrl_configs::NTuple{M, Int}) where {N, M}
step_1 = 1 << (first(locs) - 1)
step_2 = 1 << first(locs)
ctrl_mask = bmask(ctrl_locs)
flag_mask = reduce(+, 1 << (ctrl_locs[i] - 1) for i in 1:length(ctrl_locs) if ctrl_configs[i] == 1)
U11 = U[1, 1]; U12 = U[1, 2];
U21 = U[2, 1]; U22 = U[2, 2];
@inbounds for j in 0:step_2:size(st, 1)-step_1
for i in j:j+step_1-1
if ismatch(i, ctrl_mask, flag_mask)
ST1 = U11 * st[i+1] + U12 * st[i + step_1 + 1]
ST2 = U21 * st[i+1] + U22 * st[i + step_1 + 1]
st[i + 1] = ST1
st[i + step_1 + 1] = ST2
end
end
end
return st
end
let’s now compare the performance
julia> U = rand(ComplexF64, 2, 2);
julia> locs = (3, );
julia> ctrl = (4, 5);
julia> flag = (1, 1);
julia> st = rand(ComplexF64, 1<<15);
julia> @benchmark broutine!(r, $U, $locs, $ctrl, $flag) setup=(r=copy($st))
BenchmarkTools.Trial:
memory estimate: 736 bytes
allocs estimate: 10
--------------
minimum time: 17.380 μs (0.00% GC)
median time: 23.989 μs (0.00% GC)
mean time: 23.719 μs (0.00% GC)
maximum time: 46.799 μs (0.00% GC)
--------------
samples: 10000
evals/sample: 1
julia> @benchmark broutine2x2!(r, $U, $locs, $ctrl, $flag) setup=(r=copy($st))
BenchmarkTools.Trial:
memory estimate: 80 bytes
allocs estimate: 3
--------------
minimum time: 8.283 μs (0.00% GC)
median time: 8.423 μs (0.00% GC)
mean time: 8.479 μs (0.00% GC)
maximum time: 15.943 μs (0.00% GC)
--------------
samples: 10000
evals/sample: 3
Now the controlled single qubit gate routine is also improved a lot! Let’s dispatch to this too!
function broutine!(st::AbstractVector, U::AbstractMatrix, locs::NTuple{N, Int}, ctrl_locs::NTuple{M, Int}, ctrl_configs::NTuple{M, Int}) where {N, M}
size(U, 1) == 2 && return broutine2x2!(st, U, locs, ctrl_locs, ctrl_configs)
n = log2dim1(st)
subspace = bsubspace(n, sort([locs..., ctrl_locs...]))
comspace = bcomspace(n, locs)
offset = ctrl_offset(ctrl_locs, ctrl_configs)
subspace_mul!(st, comspace, U, subspace, offset)
return st
end
Parallelize using Multi-threads
Now since we have implemented general matrix instructions, we should be able to simulate arbitrary quantum circuit. We can now parallel what we have implemented using multi-thread directly as we mentioned at the beginning. However, multi-threading is not always beneficial, it has a small overhead. Thus we may not want it when the number of qubits is not large enough.
We will implement a @_threads macro as following
macro _threads(ex)
return quote
if (Threads.nthreads() > 1) && (length(st) > 4096)
$(Expr(:macrocall, Expr(:(.), :Threads, QuoteNode(Symbol("@threads"))), __source__, ex))
else
$ex
end
end |> esc
end
Parallelize using CUDA
Now, we have implemented Pauli gates and a general matrix instructions. Let’s parallelize them using CUDA.jl. Since we are not using general purpose matrix multiplication anymore, we need to write our own CUDA kernels, but this is actually not very hard in Julia, because we can reuse a lot code from our previous implementation.
But before we start doing this, let me explain what is a kernel function in the context of CUDA programming. As you might have known, GPU devices are special chip designed for executing a lot similar tasks in parallel. These tasks can be described via a function. Executing the kernel function on GPU is in equivalent to execute this function on CPU within a huge loop.
So as you have realized, this kernel function is exactly the same thing we unrolled in previous implementation. Thus we can quickly turn out previous CPU implementation into GPU implementation by wrapping the kernel into a closure, which is very mechanical. Although, the best way to do this is to move the overlapping part into a function, to demonstrate things more clearly in the blog post I just simply copy paste the previous implementation.
function broutine!(st::CuVector{T}, U::AbstractMatrix, locs::Tuple{Int}) where T
U11 = U[1, 1]; U12 = U[1, 2];
U21 = U[2, 1]; U22 = U[2, 2];
step_1 = 1 << (first(locs) - 1)
step_2 = 1 << first(locs)
function kernel(st)
idx = (blockIdx().x - 1) * blockDim().x + threadIdx().x
j = step_2 * idx - step_2
for i in j+1:j+step_1
ST1 = U11 * st[i] + U12 * st[i + step_1]
ST2 = U21 * st[i] + U22 * st[i + step_1]
st[i] = ST1
st[i + step_1] = ST2
end
return
end
N = length(0:step_2:size(st, 1)-step_1)
nblocks = ceil(Int, N/256)
@cuda threads=256 blocks=nblocks kernel(st)
return st
end
function broutine!(st::CuVector{T}, U::AbstractMatrix, locs::Tuple{Int}, ctrl_locs::NTuple{M, Int}, ctrl_configs::NTuple{M, Int}) where {T, M}
step_1 = 1 << (first(locs) - 1)
step_2 = 1 << first(locs)
ctrl_mask = bmask(ctrl_locs)
flag_mask = reduce(+, 1 << (ctrl_locs[i] - 1) for i in 1:length(ctrl_locs) if ctrl_configs[i] == 1)
U11 = U[1, 1]; U12 = U[1, 2];
U21 = U[2, 1]; U22 = U[2, 2];
function kernel(st)
idx = (blockIdx().x - 1) * blockDim().x + threadIdx().x
j = step_2 * idx - step_2
for i in j:j+step_1-1
if ismatch(i, ctrl_mask, flag_mask)
ST1 = U11 * st[i+1] + U12 * st[i + step_1 + 1]
ST2 = U21 * st[i+1] + U22 * st[i + step_1 + 1]
st[i + 1] = ST1
st[i + step_1 + 1] = ST2
end
end
return
end
N = length(0:step_2:size(st, 1)-step_1)
nblocks = ceil(Int, N/256)
@cuda threads=256 blocks=nblocks kernel(st)
return st
end
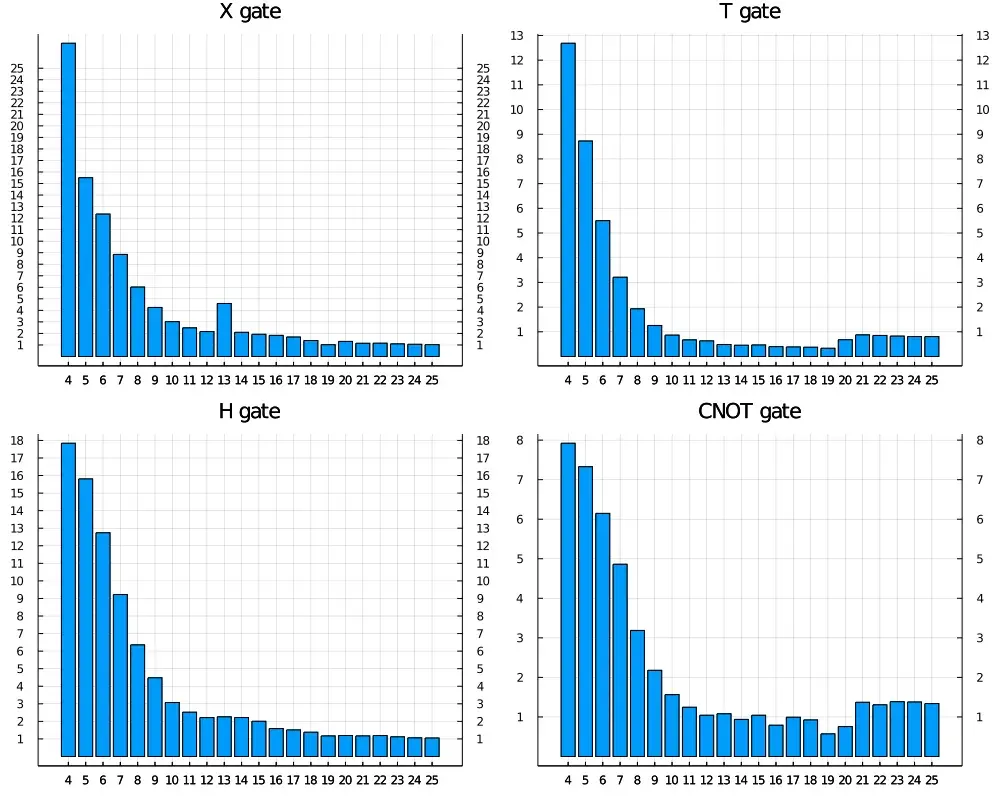
Benchmark
Now let’s see how fast is our ~600 line of code quantum circuit emulator. I don’t intend to go through a complete benchmark here
since the above implementation is generic it will has similar benchmark on different kinds of gates. And there are still plenty
of room to optimize, e.g we can specialize each routine for a known gate, such X gate, H gate to make use of their matrix structure.
The benchmark of multi-threaded routines and CUDA is currently missing since I don’t have access to a
GPU with ComplexF64 support to make the comparison fair. However, this blog post is a simple version of
YaoArrayRegister
in the Yao ecosystem, you can use the benchmark of Yao for reference. Or please also feel free to
benchmark the implementation and play with it in this blog post yourself for sure!
Let me compare this with one of the current best performance simulator qulacs, you should be able to find relative benchmark comparing qulacs and other software here. (I’m not comparing with Yao because the implementation is similar to what is implemented in Yao.)
first we clone the benchmark repo
git clone https://github.com/Roger-luo/quantum-benchmarks.git
then checkout to the stable release branch release-0.1
cd quantum-benchmarks && git checkout release-0.1
bin/benchmark setup qulacs
bin/benchmark run qulacs
this will prepare us the benchmark data on our machine. then we benchmark our own implementation
using BenchmarkTools
data = Dict(
"X" => [],
"T" => [],
"H" => [],
"CNOT" => [],
)
for n in 4:25
st = rand(ComplexF64, 1<<n)
t = @benchmark broutine!(r, $([0 1;1 0]), (3, )) setup=(r=copy($st))
push!(data["X"], minimum(t).time)
end
for n in 4:25
st = rand(ComplexF64, 1<<n)
t = @benchmark broutine!(r, $([1 0;0 exp(im * π / 4)]), (3, )) setup=(r=copy($st))
push!(data["T"], minimum(t).time)
end
for n in 4:25
st = rand(ComplexF64, 1<<n)
t = @benchmark broutine!(r, $([1/sqrt(2) 1/sqrt(2); 1/sqrt(2) -1/sqrt(2)]), (3, )) setup=(r=copy($st))
push!(data["H"], minimum(t).time)
end
for n in 4:25
st = rand(ComplexF64, 1<<n)
t = @benchmark broutine!(r, $([0 1;1 0]), (2, ), (3, ), (1, )) setup=(r=copy($st))
push!(data["X"], minimum(t).time)
end
note: we always use minimum time as a stable estimator for benchmarks
now we plot the benchmark of X, H, T, CNOT in relative time, to see how good our own simulator is comparing to
one of the best Python/C++ based circuit simulator in single thread.
What’s more?
Recall our previous implementation, since we didn’t specify our matrix type or vector type
to be a Vector or other concrete type, and didn’t specify the element type has to be a ComplexF64 either,
this means ANY subtype of AbstractVector, and ANY subtype of Number can be used with the above methods.
Now we can do something interesting, e.g we can automatically get the ability of symbolic calculation by
feeding symbolic number type from SymEngine package or SymbolicUtils package.
Or we can use Dual number to perform forward mode differentiation directly. Or we can estimate error
by using the error numbers from Measurements.
Here is demo of using SymEngine:
using SymEngine
julia> @vars α θ
(α, θ)
julia> st = Basic[1, α, 0, 0]
4-element Array{Basic,1}:
1
α
0
0
julia> broutine!(st, [exp(-im * θ) 0; 0 exp(im * θ)], (1, ))
4-element Array{Basic,1}:
exp(-im*θ)
exp(im*θ)*α
0
0
This is only possible when one is able to use generic programming to write high performance program, which is usually not possible in the two-language solution Python/C++ without implementing one’s own type system and domain specific language (DSL) compiler, which eventually becomes some efforts that reinventing the wheels.
Conclusion
Getting similar performance or beyond comparing to Python/C++ solution in numerical computation is easily achievable in pure Julia with much less code. Although, we should wrap some of the overlapping code into functions and call them as a better practice, we still only use less than 600 lines of code with copy pasting everywhere.
Moreover, the power of generic programming will unleash our thinking of numerical methods on many different numerical types.
Experienced readers may find there may still rooms for further optimization, e.g we didn’t specialize much common gates yet, and the loop unroll size might not be the perfect size, and may still vary due to the machine.
Last, besides simulating quantum circuits, the above implementation of subspace matrix multiplication is actually a quite common routine happens frequently in tensor contraction (because quantum circuits are one kind of tensor network), thus more promising application can be using these routines for tensor contraction, however, to make these type of operations more efficient, it may require us to implement BLAS level 3 operation in the subspace which is the subspace matrix-matrix multiplication, which can require more tricks and more interesting.
I uploaded the implementation as a gist: https://gist.github.com/Roger-luo/0df73cabf4c91f9854657fdd2ed66767